UPDATE: 2021-05-13: zoomGroupStats is now available as a package on CRAN.
# To use the stable release version of zoomGroupStats, use:
install.packages("zoomGroupStats")
# To use the development version, which will be regularly updated with new functionality, use:
library(devtools)
install_github("https://github.com/andrewpknight/zoomGroupStats")
You can stay up-to-date on the latest functionality on http://zoomgroupstats.org.
UPDATE: 2021-04-30: In response to prodding from several folks who have been using the functions, I have started to build these R functions for analyzing Zoom meetings as a package. I’m grateful for all of the feedback and suggestions for features and modifications to this project. Although things are still in flux, you can now access a package version of zoomGroupStats. The easiest way is to use the dev_tools package and install the current development version from my github repository. To do so, you can run:
library(devtools)
install_github("https://github.com/andrewpknight/zoomGroupStats", force=TRUE)
library(zoomGroupStats)
I have created a multi-part guide for using this package to conduct research using Zoom and analyze data from Zoom, which I will continue to extend and elaborate. To keep up-to-date on this work, please use the dedicated package website http://zoomgroupstats.org.
Please keep the feedback and comments coming!
UPDATE: 2021-02-12: I’ve updated several items in the package. I’m also going to moving the development and updates all over to github to ease version control and documentation. For now, here is a post that is from a recent live tutorial session I facilitated. Stay tuned!
UPDATE: 2020-07-27: The new file contains some alpha-stage functions for doing audio transcription and for conducting a windowed conversation analysis. I haven’t yet tested these functions extensively or commented the windowed conversation analysis. Once COVID teaching planning eases, I’ll get back in an update further.
UPDATE: 2020-04-14: I added a new function (textConversationAnalysis) that gives some very basic and descriptive conversation metrics from either the video transcript or the chat file.
You can always access the most recent version of the functions by including the statement source(“http://apknight.org/zoomGroupStats.R”) at the top of your code.
Alternatively, you could go to http://apknight.org/zoomGroupStats.R and copy/paste the code into your editor.
ORIGINAL POST FOLLOWS:
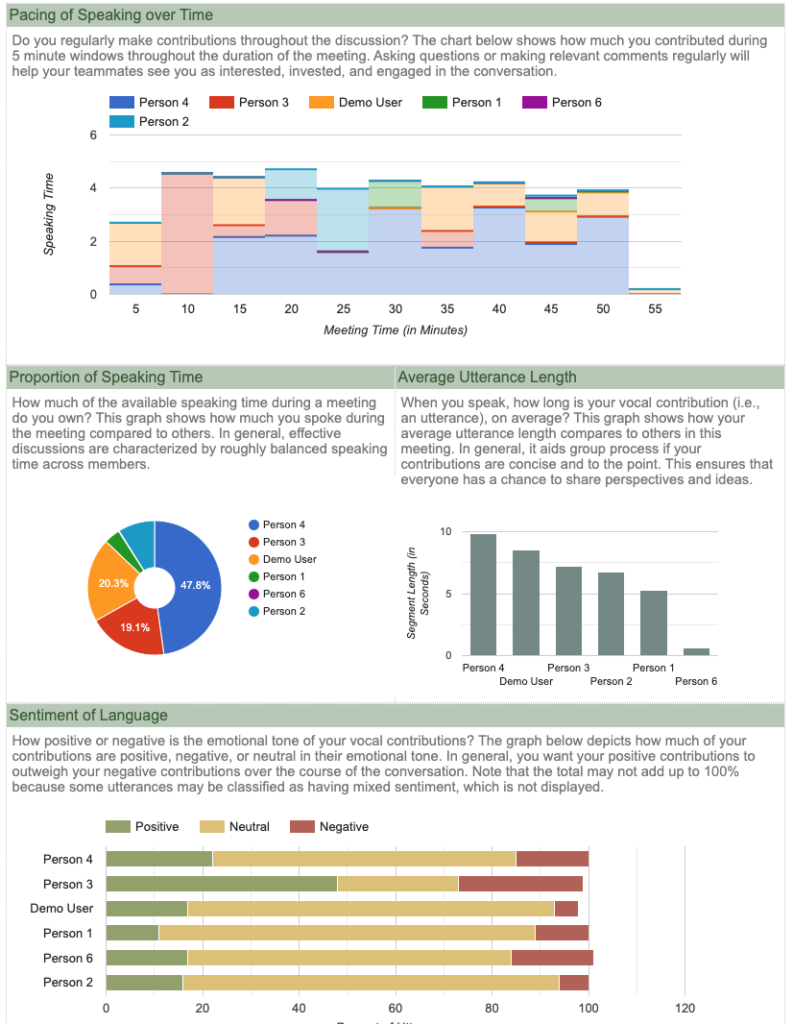
In response to the shift to so many online meetings, I created a set of R functions to help do research using web-based meetings. In brief, these functions use the output of recorded sessions (e.g., video feed, transcript file, chat file) to do things like sentiment analysis, face analysis, and emotional expression analysis. In the coming week, I will extend these to do basic conversation analysis (e.g., who speaks/chats most, turntaking).
I went overboard in commenting the code so that hopefully others can use them. But, if you’re still having trouble getting them to work, please don’t hesitate to reach out to me.
You can directly access the functions here:
http://apknight.org/zoomGroupStats.R
After reviewing this, you could call these functions using the following statement in R: source(“http://apknight.org/zoomGroupStats.R”)